(I apologise in advance for not staying on topic long enough to discuss matters in more detail. This is largely due to my fleeting attention span at the time of writing and looming deadlines IRL. I hope you still like this relatively short post!)
This piece of graffiti I photographed whilst taking a stroll down Regent’s Canal reminded me of the secondary structure of proteins:
No magic eye images here thankfully, just an alpha-helix and two unstructured loops either side of it that have cyclized. I’m sure there are real-life examples of simple, short peptides like this. “What is the smallest structured protein?”, I hear you ask? Why that is a very good question….a question I will probably cover in more detail at a later date.
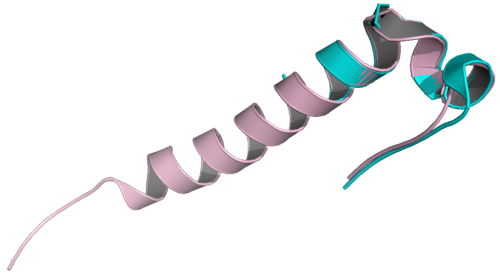
As a quick teaser (yeah right), here is the TRP-Cage miniprotein:
This protein is composed of 20 amino acids and was truncated and mutated from a larger (39 amino acids) protein, Exendin-4, orginally isolated from the oral secretions of the Gila Monster. The authors solved the structures of both exendin-4 and the TRP-Cage miniprotein by solution-state NMR. The two share remarkable structural homology, as can be seen when they are superimposed:
You can find the original papers describing these two proteins here and here.
It was during an external seminar back in my undergrad days that I first heard about designing proteins from scratch (de novo protein design) and the field of protein engineering. The field is part of the larger, more well known field of synthetic biology (loads of resources on the subject can be found here), but has found use in attempting to answer more basic research questions such as those in protein folding.
The idea of taking a protein out of nature, fiddling around with it, fusing it to another protein, mutating a few regions to alter binding ability or enzyme specificity is one that continues to fascinate me. We are now reaching the point in our collective understanding where we know enough about a given system or molecules to begin dismantling discrete portions and putting them back together in new ways to produce something with a curiously awesome function. The potential benefits and solutions such a discipline could bring to today’s problems is near limitless. I will most definitely expand on this in a later post.
Thanks for reading, and if by chance you know of any other small, folded proteins that can be mentioned in my eventual, longer post, please let me know in the comments section! You can also give your own input, including constructive criticism, in the comments section, and maybe get a discussion going.
Until next time, happy protein designing!